Research
Overview
With unprecedented growth in computationally intensive areas using large datasets from novel sources, the use of statistics is expanding rapidly inside and outside of academia. How to efficiently and accurately decipher complex relationships in large-scale data from real-world problems remains an urgent challenge. In recent years, a confluence of advances in machine learning and statistics is fueling a new wave of innovation that is poised to improve data aggregation, estimation, interpretation, and prediction. Many classical disciplines of statistics are being redefined and reinforced by the magnificent bloom of machine learning and artificial intelligence. My general research interest lies in this multi-disciplinary area where I have been committed to developing practical statistical and machine learning tools with significance in both statistical theory and applications. In particular, I have been pursuing this research agenda by exploiting the advances in generative artificial intelligence (AI) to tackle several fundamental statistical problems, such as density estimation, causal inference, and unsupervised learning with broad applications in computational biology and biomedical informatics.
My research goal is to fulfill the new theories and methodologies for solving both statistical problems and data science problems in computational biology by developing computationally efficient techniques from machine learning and AI field. I have been developing novel frameworks for causal inference (arXiv 2022), density estimation (PNAS 2021), using generative AI and apply the relative techniques to various computational biology problems, which include genomic studies (NMI 2021, BIB 2022,ISMB/Bioinformatics 2019), and pharmacology studies (ECCB/Bioinformatcis 2020,NeurIPS 2020).
Representative works in Statistics
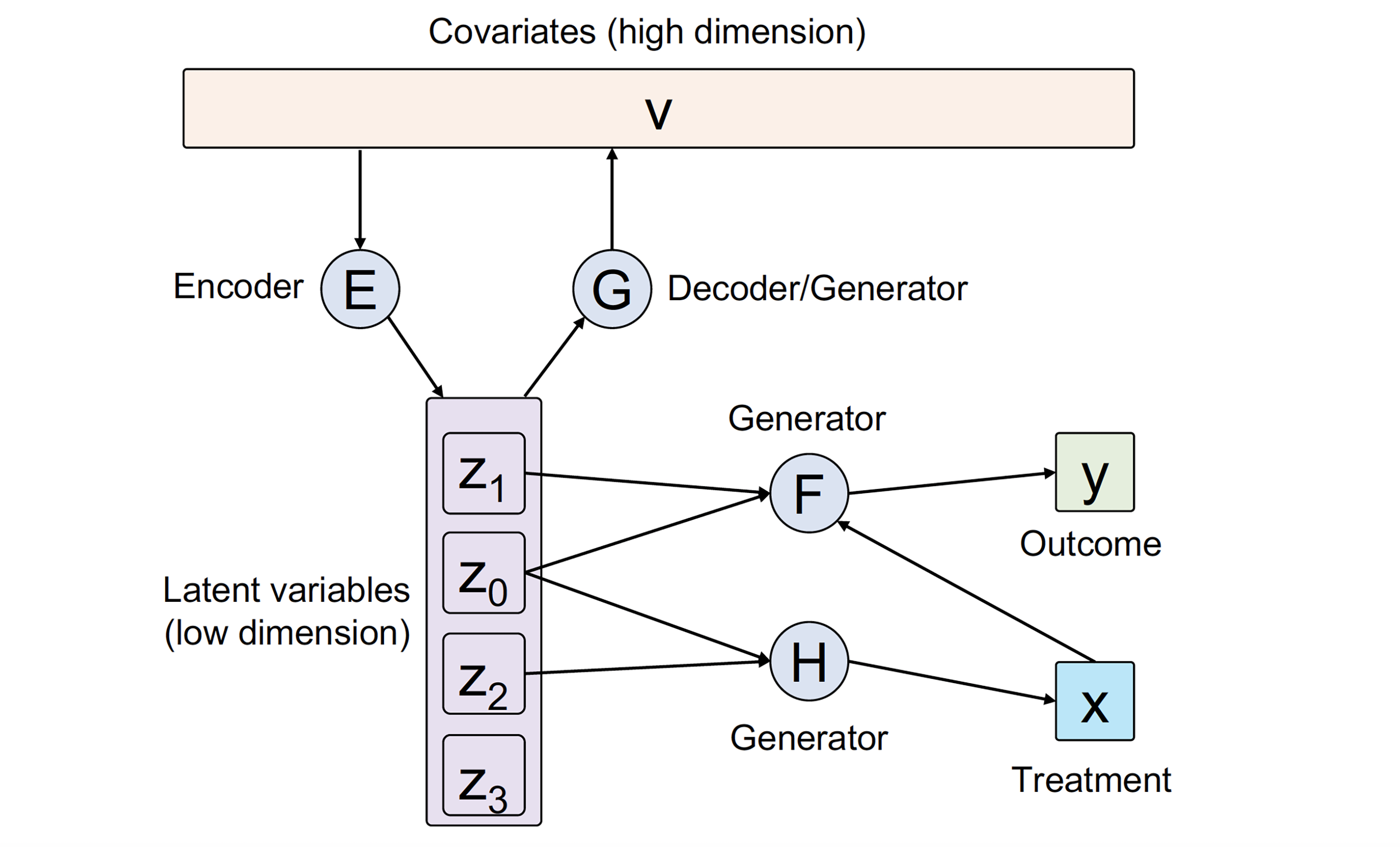
In this article, we develop a general framework CausalEGM for estimating causal effects by encoding generative modeling, which can be applied in both binary and continuous treatment settings. We establish a bidirectional transformation between the high-dimensional confounders space and a low-dimensional latent space where the density is known. Through this, CausalEGM simultaneously decouples the dependencies of confounders on both treatment and outcome and maps the confounders to the low-dimensional latent space. By conditioning on the low-dimensional latent features, CausalEGM can estimate the causal effect for each individual or the average causal effect within a population. Our theoretical analysis shows that the excess risk for CausalEGM can be bounded through empirical process theory. Under an assumption on encoder-decoder networks, the consistency of the estimate can be guaranteed.
Qiao Liu, Zhongren Chen, Wing Hung Wong
arXiv preprint arXiv:2212.05925, 2022
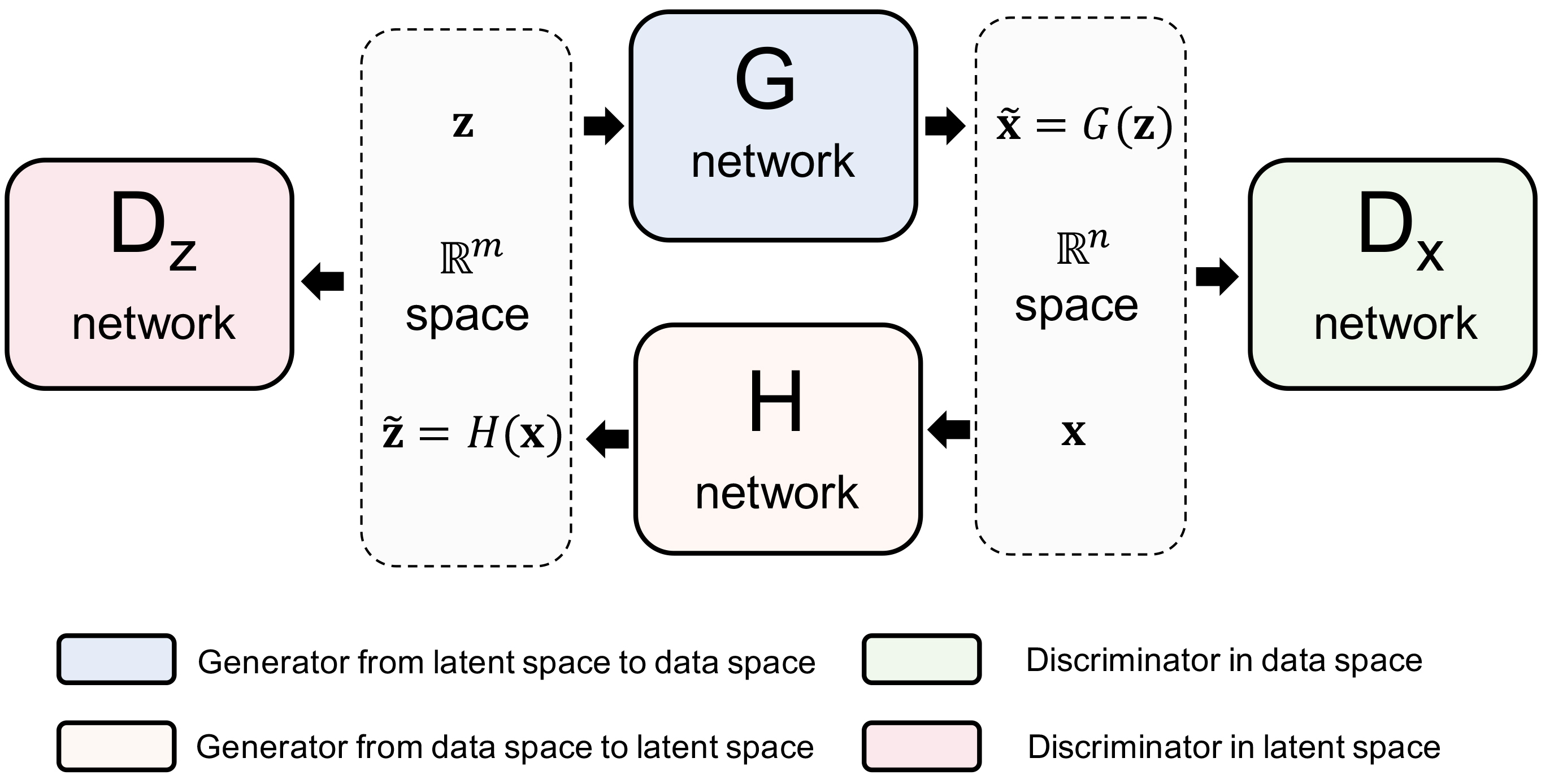
Density estimation is among the most fundamental problems in statistics. It is notoriously difficult to estimate the density of high-dimensional data due to the “curse of dimensionality”. Here, we introduce a new general-purpose density estimator based on deep generative neural networks. By modeling data normally distributed around a manifold of reduced dimension, we show how the power of bidirectional deep generative models can be exploited for explicit evaluation of the data density by either importance sampling or Laplacian approximation. Simulation and real data experiments suggest that our method is effective in a wide range of problems. This approach should be helpful in many applications where an accurate density estimator is needed.
Qiao Liu, Jiaze Xu, Rui Jiang, Wing Hung Wong
Proceedings of the National Academy of Sciences (PNAS), 2021, 18(15):e2101344118
Representative works in Computational Biology
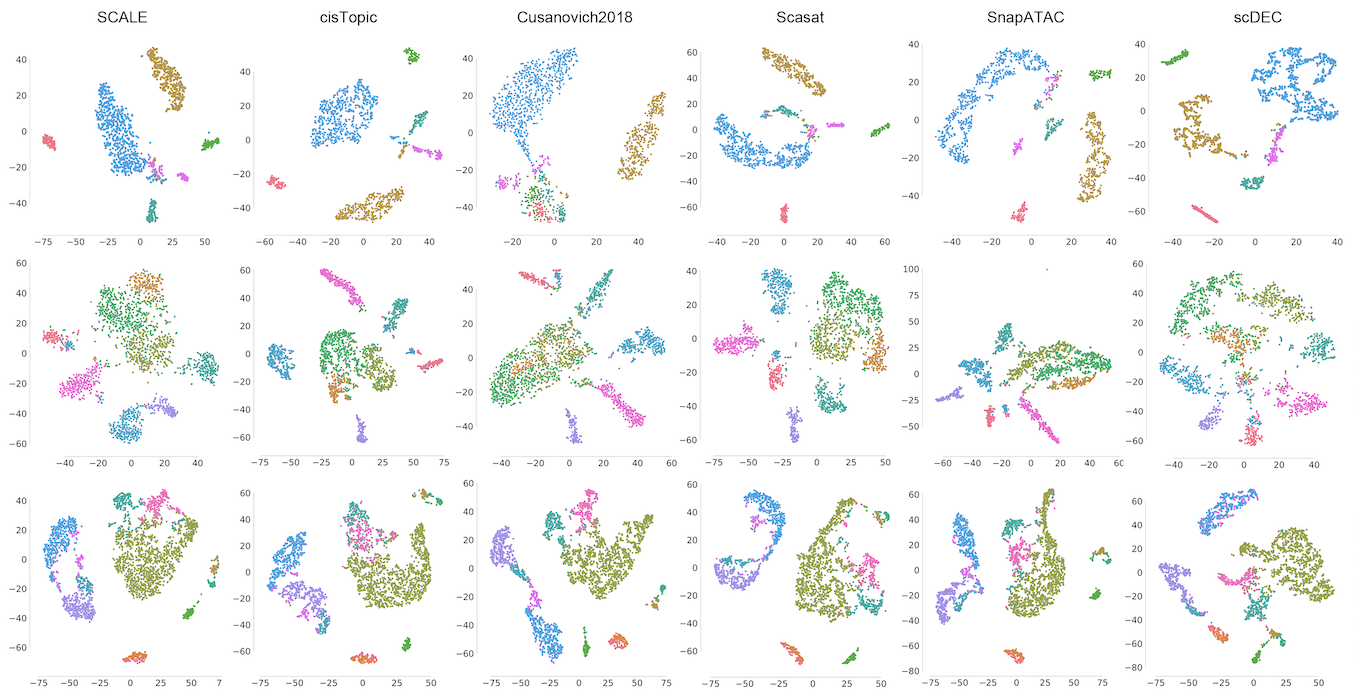
We proposed scDEC, a computational tool for single cell data analysis with deep generative neural networks. scDEC is built on a pair of generative adversarial networks (GANs), and is capable of learning the latent representation and inferring the cell labels, simultaneously. scDEC can be used for clustering scATAC-seq and Multiome data.
Qiao Liu, Shengquan Chen, Rui Jiang, Wing Hung Wong
Nature Machine Intelligence, 2021, 3(6):536-544.
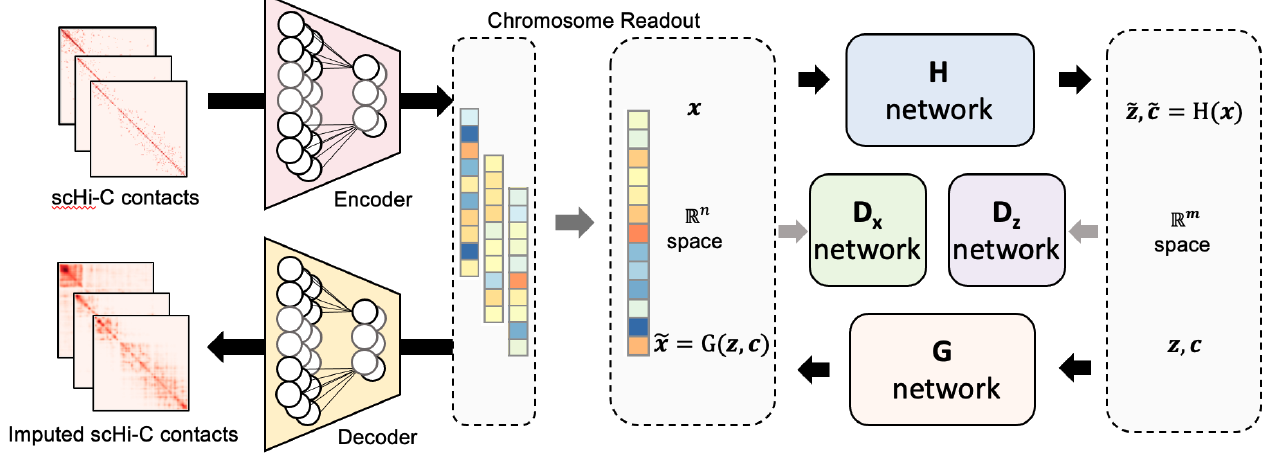
Deciphering 3D genome conformation is important for understanding gene regulation and cellular function at a spatial level. Here, we proposed scDEC-Hi-C, a new framework for single cell Hi-C analysis with deep generative neural networks.
Qiao Liu, Wanwen Zeng, Wei Zhang, Sicheng Wang, Hongyang Chen, Rui Jiang, Mu Zhou, Shaoting Zhang
Briefings in Bioinformatics, 2022.
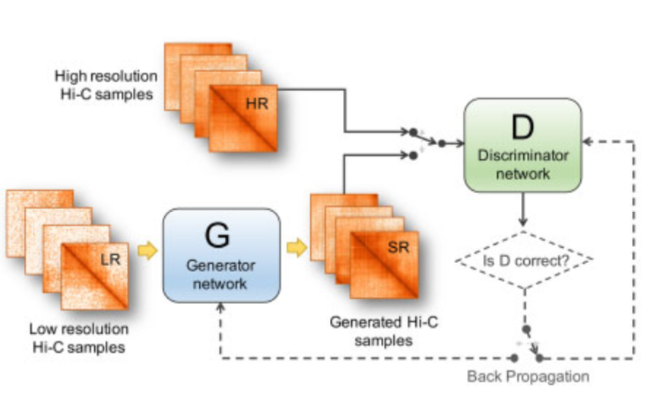
We proposed hicGAN for inferring high resolution Hi-C data from low resolution Hi-C data with generative adversarial networks (GANs). To the best of our knowledge, this is the first study to apply GANs to 3D genome analysis. Pretrained hicGAN models were also provided.
Qiao Liu, Hairong Lv, Rui Jiang
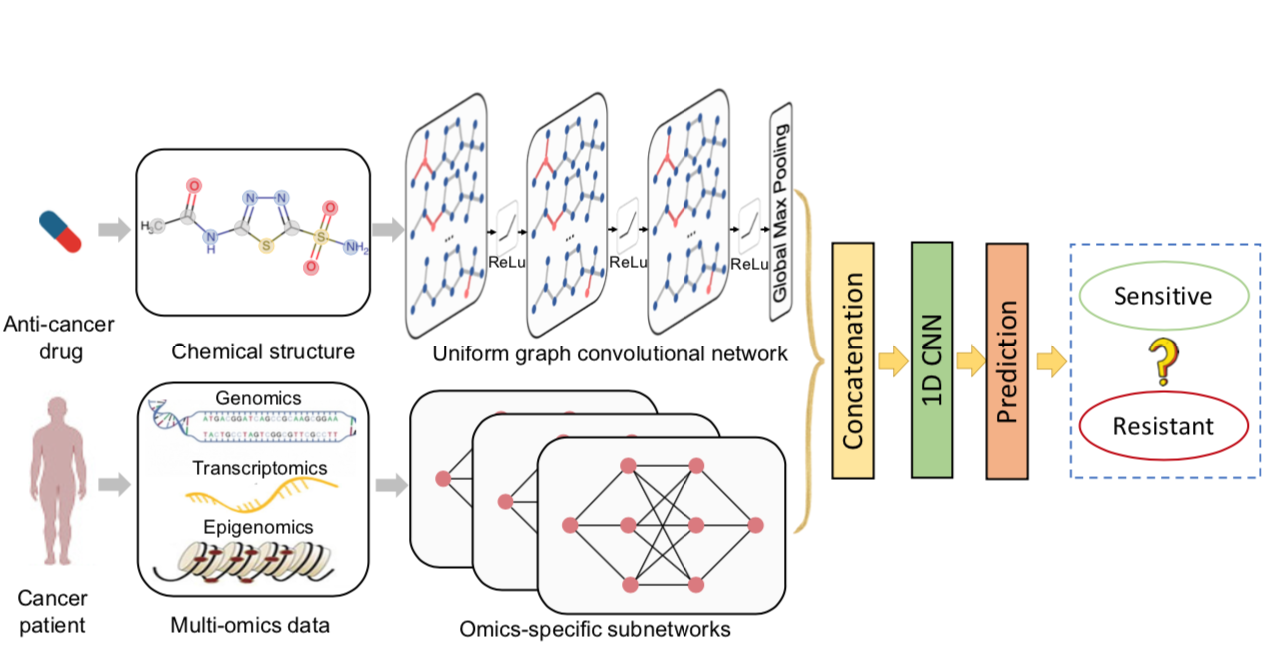
DeepCDR is a hybrid graph convolutional network consisting of a uniform graph convolutional network (UGCN) and multiple subnetworks. Unlike prior studies modeling hand-crafted features of drugs, DeepCDR automatically learns the latent representation of topological structures among atoms and bonds of drugs. DeepCDR achieves state-of-the-art performance in cancer drug response prediction.
Qiao Liu, Zhiqiang Hu, Rui Jiang and Mu Zhou
